The third, and most important, kind of R object we will study is called a dataframe, which is how R typically stores statistical data. Although you can make them on your own with the data.frame() function a lot of them come built in or can be downloaded as needed.
To start we’ll make our own dataframe using the results of the 2014 Boston Marathon Wheelchair Division. The data is made up of several vectors. The placement of the racer, their sex, the time (in minutes) they took to finish, and the country they raced for. Feel free to copy and paste these rather than type them out by hand.
Place <- c(1:5,1:5)
Sex <- c(rep("Male",5),rep("Female",5))
Time <- c(80.60,81.23,81.23,84.65,84.70,95.10,97.40,98.55,99.65,101.70)
Country <- c("RSA","JPN","JPN","SUI","ESP","USA","JPN","USA","SUI","GBR")
Now we use the data.frame() function to combine them.
boston <- data.frame(Place,Sex,Time,Country)
boston
Place Sex Time Country
1 1 Male 80.60 RSA
2 2 Male 81.23 JPN
3 3 Male 81.23 JPN
4 4 Male 84.65 SUI
5 5 Male 84.70 ESP
6 1 Female 95.10 USA
7 2 Female 97.40 JPN
8 3 Female 98.55 USA
9 4 Female 99.65 SUI
10 5 Female 101.70 GBR
Short and easy to understand. Each line represents a single person. Line 8 is the third place woman she was an American who took 98.55 minutes to complete the race. You may notice that a dataframe is somewhat like a matrix. The largest difference is in how R treats them, a number of functions work especially well with dataframes. Since we have this convenient dataframe to work with let’s take the opportunity to learn about structure with the str() function.
str(boston)
'data.frame': 10 obs. of 4 variables:
$ Place : int 1 2 3 4 5 1 2 3 4 5
$ Sex : Factor w/ 2 levels "Female","Male": 2 2 2 2 2 1 1 1 1 1
$ Time : num 80.6 81.2 81.2 84.7 84.7 ...
$ Country: Factor w/ 6 levels "ESP","GBR","JPN",..: 4 3 3 5 1 6 3 6 5 2
Race results are good because they include many different kinds of data and this gives us a chance to see how R treats them. We’ll talk about levels of measurement in a later post but for now notice the information that str() gives you about the data.
There are “10 obs. of 4 variables” which means the dataframe is ten lines long and has four pieces of information (Place, Sex, Time, and Country). Next it tells you about each variable. The Place variable is made of integers, whole numbers only. The Sex variable is a “Factor w/ 2 levels” which means the information does not have a numerical form and there are two options. The Time variable is a numeric which means it can hold any numerical value, including decimals. The Country variable is a “Factor w/ 6 levels” which means that, like Sex, it doesn’t have numerical form and there are six options.
Since the boston data is extremely small you could have figured all of this out by yourself but in practice dataframe can be thousands of lines long and have many variables. With the str() function you can get a simple explanation of all of them.
The subsetting notation we saw before with vectors and matrices also works with dataframes. For example let’s look at row seven.
boston[7,]
Place Sex Time Country
7 2 Female 97.4 JPN
All of the columns in dataframes are variables with names and names are easier to remember than the column numbers so R allows you to extract a variable by using its name. To extract the variables you can use $ notation as well as the [ ] notation. These commands are all equivalent.
boston$Time
boston[,3]
boston[,"Time"]
Now that we have a dataframe to play with we can look at more complex subsetting commands that are frequently useful. Let’s look at a few of them. See if you can follow along, we’re just combining commands we’ve seen before, essentially making subsets of subsets.
Only the Japanese racers:
boston[boston$Country=="JPN",]
Place Sex Time Country
2 2 Male 81.23 JPN
3 3 Male 81.23 JPN
7 2 Female 97.40 JPN
Only the woman in first place:
boston[boston$Sex=="Female" & boston$Place==1,]
Place Sex Time Country
6 1 Female 95.1 USA
It’s also possible to apply functions to subsets. For example let’s look at the average of the times and then average for each of men and women by using the mean() function on different pieces of the dataframe.
mean(boston$Time)
90.481
mean(boston$Time[boston$Sex=="Male"])
82.482
mean(boston$Time[boston$Sex=="Female"])
98.48
The boston data is extremely small and simple. You could probably understand everything you want to know about it just by looking at the whole thing. In practice you’ll want to use R with data that is significantly larger and more complicated. The Old Faithful data is one of the most frequently analyzed data sets in all of statistics. It comes built into R as the faithful dataframe which you can call up any time you want.
Because it is much larger than boston we should start with the str() function in order to understand it.
str(faithful)
'data.frame': 272 obs. of 2 variables:
$ eruptions: num 3.6 1.8 3.33 2.28 4.53 ...
$ waiting : num 79 54 74 62 85 55 88 85 51 85 ...
That’s actually smaller than the structure of the boston data because there are only two variables but take a look at that first line. Yep, faithful has 272 rows! That’s much too large for us to try and understand just by looking at the whole dataframe. Read the descriptions of that eruptions variable and the waiting variable to see what they tell you.
Since faithful is built into R you can get detailed information from the internet by using the help function.
?faithful
That webpage tells us that the eruptions variable is the length of the eruption in minutes and the waiting variable is the delay between that eruption and the next. The task of a data scientist is to transform data into information, so that we can eventually gain knowledge. There are countless ways of getting information out of the data. One of the most popular is to calculate summary statistics. The summary() function computes several popular ones, all of which can be calculated individual if you’d prefer.
summary(faithful)
eruptions waiting
Min. :1.600 Min. :43.0
1st Qu.:2.163 1st Qu.:58.0
Median :4.000 Median :76.0
Mean :3.488 Mean :70.9
3rd Qu.:4.454 3rd Qu.:82.0
Max. :5.100 Max. :96.0
The min() and max() are the smallest and largest values. The mean() is the average value. The median() is the central value. The 1st and 3rd quartiles mark the bottom 25% and top 25% of the data, exactly half of the data falls between those values.
We can see here that old faithful isn’t quite as faithful as you may have been lead to believe. The eruptions range from 1.6 to 5.1 minutes in duration and occur between 43 and 96 minutes apart. Even if we look at the quartiles, to ignore outliers, there is a significant degree of variability.
There is a better way. Watch this:
plot(faithful)
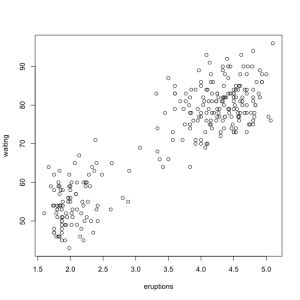
The ease with which basic graphics can be made is one of the best things about R. More complex visualizations are better done with one of the many graphics packages available but base R graphics can efficiently do a lot of work. Looking at the plot we see that there are basically two kinds of eruptions, long ones that are followed by a long delay and short ones that are followed by a short delay.
The additional drawing functions let us quickly add things to the graphics we make. Let’s draw a line dividing the two groups. Crudely we might draw it at the mean of the eruption duration. While the scatterplot is open use the abline() function to draw a straight vertical line.
abline(v=mean(faithful$eruptions))
Finally lets look at a smaller but more complex dataframe called mtcars. It is only 32 lines long but each type of car was measured for 11 variables. You’ve made use of the srt() and plot() functions before so try theme with mtcars on your own. See what conclusions you can draw then using the ? function to learn more about it.
When you tried the plot() function you probably got a huge grid of scatterplots. This scatterplot matrix can be intimidating but it really isn’t that different from a normal scatterplot. Let’s use the subsetting notation to look at just a few of the variables.
plot(mtcars[,c("mpg","disp","hp","drat","wt","qsec")])
Take a second to examine this line of code, you should already be familiar with each part of it. We use the plot() function on a subset of mtcars that we define with a vector made up of strings, thanks to the c() function we learned about earlier.
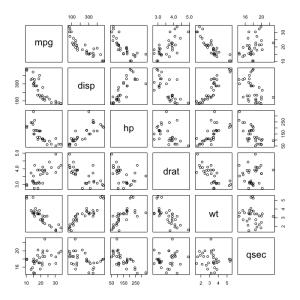
To read a plot matrix find the place where the two variables you’re interested in intersect. For example immediately below the mpg (miles per gallon) cell is the scatterplot showing its relationship to the disp (displacement or volume) of the car. It looks to me like as the size of the car increases mileage decreases. The same relationship looks to be true of mpg and hp (horsepower) while the opposite is true of wt (weight) an disp.
In the next post we’ll take a look at solving and visualizing a simple problem in R as a way to practice working with data.