In our last post we addressed the question of where exactly the t-distribution used in a t-test comes from.
A t-test is probably the most frequently used statistical test in modern science. It works almost identically to the z-Test (and follows the same concept as the binomial test except) that we are using the more arcane t-distribution to describe a sampling distribution.
Consider the information we have about the weight of baseball players. The weight of those 1033 players is 201 pounds. On the Arizona team, however, the average weight is 208 pounds.
Here is the mean and SD for that team.
mean(tab$weight[tab$team == "ARZ"]) 208.0714 sd(tab$weight[tab$team == "ARZ"]) 24.5386
How do we test to see if this difference is likely to occur by chance than deliberate choice?
Our hypothesis is that the Arizona players are not being picked for their weight so their drawn from a population with a mean of 200 pounds, like everybody else. To find the t-value we subtract the hypothetical mean from sample mean and divide by the sample standard deviation, this is the equation derived last time. We’re attempting to normalize the errors.
arz <- tab$weight[tab$team == "ARZ"] m.pop <- mean(tab$weight) m.arz <- mean(arz) s.arz <- sd(arz)/sqrt(length(arz)) t <- (m.arz-m.pop)/s.arz t 1.376252
This is a value that we compare to the t-distribution!
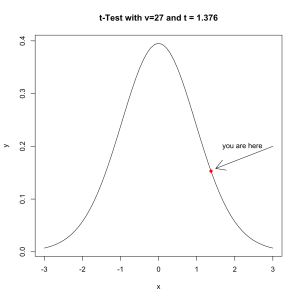
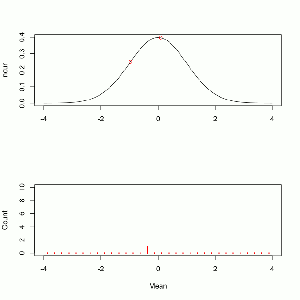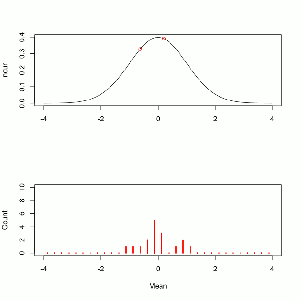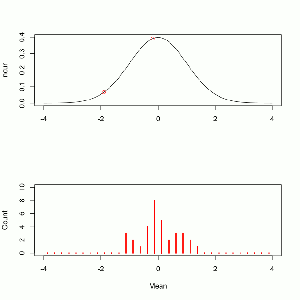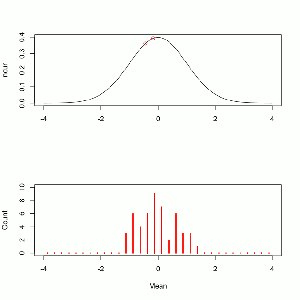
Since there are 28 players on the team we use a distribution with a ν of 27. The density looks like this.
x <- seq(-3,3,0.01) y <- dt(comb,27) plot(y~x,type='l', main='t-Test with v=27 and t = 1.376') points(dt(t,27)~t,pch=16,col='red') arrows(3,.2,1.5,dt(t,27)+.005) text(2.2,.2,label='you are here')
Like with the binomial test we are looking for the probability of getting a value equal to or greater than the one we observed. The inverse cumulative tells us, for each point, the probability of selecting a greater value.
1-pt(t,27) # pt() is the cumulative t-distribution function, subtracting it from 1 produces the inverse 0.09002586
We conclude from this that, if Arizona doesn’t tend to pick players with above average weights, there is about a 9% chance of this occurring. That isn’t considered to be impressive evidence so traditionally we would not choose to draw any conclusions here.
But why should you believe me? Surely I’m just a person on the internet who could easily make a mistake while I type. Fortunately R comes with a built in t.test() function.
t.test(arz,mu=m.pop,alternative='greater')
One Sample t-test
data: arz
t = 1.3763, df = 27, p-value = 0.09003
alternative hypothesis: true mean is greater than 201.6893
95 percent confidence interval:
200.1727 Inf
sample estimates:
mean of x
208.0714
There we have it, R confirms my previous claims. The t-statistic is 1.3763, the degrees of freedom are 27, and the p-value is 0.09.