Probability Distributions
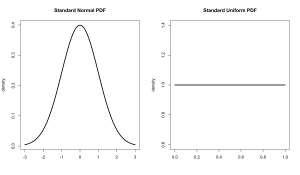
A probability density distribution is an idealized version of the kind of densities we’ve looked at many times before on this blog. Probably the most well known distributions are the uniform distribution and normal (or Gaussian) distributions. Here’s what they look like.
The standard uniform distribution is defined as having a minimum of 0 and a maximum of 1 while the standard normal distribution has a mean of 0 and a standard deviation of 1 (and stretches infinitely in both directions). Each kind of distribution has an equation that uses these characteristics to determine the density at any given point.
The uniform distribution is flat, all values are equally likely (equiprobable), while the normal distribution is bell shaped, values close to the middle are more likely than ones on the tail ends. Reading a probability density requires a small amount of abstract thinking. The density measures how dense that point is, not how probable it is. Probability is shown by area. We’ll get to that later.
It is often said that values are “drawn from” a distribution as if it were an infinitely large deck of cards. For example if you measure the weight of many adults you are drawing from an approximately normal distribution. R can draw random numbers from a lot of different distributions with the ‘r’ type functions (rnorm(), runif(), rgamma(), and so on).
The arguments for these functions require us to define the them in different ways. For the normal distribution we define the mean and and standard deviation, it will draw values in an almost infinitely wide range. For the uniform distribution we simply define the range in which we draw from, since it has no other characteristics. These values are called their parameters.
Here we draw ten values from a standard normal distribution (mean = 0, sd = 1) and from the uniform distribution between 20 and 30. The numbers are rounded off so they’re easier to read.
rnorm(10,0,1) -1.00 0.86 -0.46 1.49 -1.77 2.76 -1.48 0.23 -1.83 -0.61 runif(10,20,30) 26.82 22.77 20.23 24.53 22.79 28.88 28.58 20.38 27.10 23.43
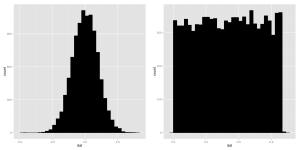
Notice that although the normal distribution could theoretically have values of any size what we find is that most of our random numbers are close to zero. The uniform numbers span the whole possible range. We can take a look at how the samples are spread out with a histogram or a density.
a <- data.frame(dat = runif(10000,0,1)) b <- data.frame(dat = rnorm(10000,0,1)) ggplot(a, aes(x=dat)) + geom_histogram(color='black',fill='black') ggplot(b, aes(x=dat)) + geom_histogram(color='black',fill='black')
Yep, they look like ragged versions of the idealized distributions they come from.
To estimate the probability of drawing a certain values from a distribution we use can the process of numeric integration to determine the area under the line (to review, we add up the density of a bunch of slices and then divide by the number of slices). What is the probability of drawing a number between -1 and 1 from a standard normal distribution? How about extremely close to the center, between -0.5 and 0.5?
# Total probability of standard normal distribution from -1 to 1. sum(dnorm(seq(-1,1,.001)))/1001 0.6822492 # Total probability of standard normal distribution from -0.5 to 0.5. sum(dnorm(seq(-0.5,0.5,.001)))/1001 0.3828941 # Total probability of standard from 0.3 to 0.72. sum(dunif(seq(0.3,0.72,.001)))/1001 0.4205794
Notice that we’re off by a bit on each of these because we’re only using 1001 slices, it might be better to use more. You can see this most obviously with the uniform distribution since it is easy to figure out the answer analytically. The probability of getting a number between 0.3 and 0.72 (for a standard uniform which, recall, goes from 0 to 1) is exactly 0.42 since it covers 42% of the range.
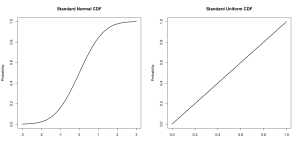
A probability distribution can be represented in a variety of ways. The cumulative distribution function, for example, shows the total probability of drawing a given value of lower. It is accessed with the ‘p’ type functions (pnorm(), punif(), pgamma(), and so on).
Notice that this is not a density function and it does show probability at each point. The function reaches 0.5 at zero for a standard normal distribution, there is a 50% chance of drawing a value lower than 0.
One last thing before we move on to the next post. For a density function, what is the probability at a given point? That is to say what is the probably of drawing a zero from the standard normal? What is the probability of drawing a three? Not an easy question. We can say that the density at zero is 0.3989 and the density at three is 0.0044 so we can say that drawing a zero is about ninety times as likely. If we squeeze our interval tighter and tighter, though, hoping to determine an exact probability both of them will drop forever toward zero. The probability is infinitesimal, effectively zero.
The purpose of this aside is to reinforce an observation that I’ve made before: It is important to think in terms of intervals rather than point values.